Introduction to eQTL analysis
Peter Humburg
Overview
- Hands-on : Setup
- What are eQTL?
- Detecting eQTL – A simple model
- Hands-on : Simple linear regression
- Detecting eQTL – Not that simple
- Hands-on : Covariates
- Covariates – Choose wisely
- If only we knew – Covariates for real data
- Hands-on : Dealing with real data
- Large scale eQTL analysis
- Hands-on : Scaling it up
- Interpreting results
- Summary
Hands-on : Setup
Docker image
Make sure you have the latest version of the docker image.
docker pull humburg/eqtl-introDirectory structure
- Create a working directory for this course. (
eqtl_course) - Within that directory, create two sub-directories
genotypesanalysis
- Mac: create directories under
/Users/ - Windows: create directories under
C:\Users\
Additional data
The docker image contains most of the data. Further genotyping data is available from
ftp://galahad.well.ox.ac.ukUsername: eqtl_course
Download the data and save it into genotypes
Start the RStudio server
Windows :
docker run -p 8787:8787
-v /c/Users/user/eqtl_course/genotpes:/data/genotypes
humburg/eqtl-introIP address of the server usually is 192.168.59.103.
Use boot2docker ip to check if necessary.
Start the RStudio server
Mac :
docker run -p 8787:8787
-v /Users/user/eqtl_course/genotpes:/data/genotypes
humburg/eqtl-introIP address of the server usually is 192.168.59.103.
Use boot2docker ip to check if necessary.
Start the RStudio server
Linux :
docker run -p 8787:8787
-v /home/user/eqtl_course/genotpes:/data/genotypes
-e USER=$USER -e USERID = $UID
humburg/eqtl-introIP address of the server usually is 127.0.0.1 (or localhost).
Using RStudio
Access the RStudio interface at http://yourip:8787.
* Username: rstudio
* Password: rstudioUseful resources
- RStudio cheat sheets
What are eQTL?
Quantitative trait loci
QTL are regions of the genome associated with quantitative traits
- height
- BMI
- lung capacity
- …
Expression quantitative trait loci
If the trait of interest is the expression of a gene, we talk about eQTL.
- Associations can be local (cis)
- or distant (trans)

Expression quantitative trait loci
If the trait of interest is the expression of a gene, we talk about eQTL.
- Associations can be local (cis)
- or distant (trans)

Expression quantitative trait loci
If the trait of interest is the expression of a gene, we talk about eQTL.
- Associations can be local (cis)
- or distant (trans)

Detecting eQTL – A simple model
How do we know whether a locus is associated with the expression of a gene?
- Any given locus contains multiple SNPs.
- Can determine genotypes for these SNPs in a (large) number of individuals.
- Measure gene expression for genes of interest.
- Assess the evidence that expression varies with genotype.
How do we know whether a locus is associated with the expression of a gene?
| sample | snp_1 | snp_2 | snp_3 | … |
|---|---|---|---|---|
| sample 1 | AA | AA | AB | … |
| sample 2 | AB | AB | AA | … |
| sample 3 | AB | BB | AB | … |
| … | … | … | … | … |
- Any given locus contains multiple SNPs.
- Can determine genotypes for these SNPs in a (large) number of individuals.
- Measure gene expression for genes of interest.
- Assess the evidence that expression varies with genotype.
How do we know whether a locus is associated with the expression of a gene?
| sample | gene_1 | gene_2 | gene_3 | … |
|---|---|---|---|---|
| sample 1 | 7.3 | 12.8 | 6.5 | … |
| sample 2 | 10.9 | 9.6 | 8.8 | … |
| sample 3 | 9.5 | 10.7 | 15.1 | … |
| … | … | … | … | … |
- Any given locus contains multiple SNPs.
- Can determine genotypes for these SNPs in a (large) number of individuals.
- Measure gene expression for genes of interest.
- Assess the evidence that expression varies with genotype.
How do we know whether a locus is associated with the expression of a gene?
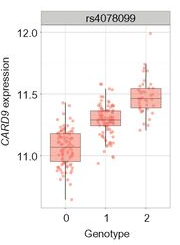
- Any given locus contains multiple SNPs.
- Can determine genotypes for these SNPs in a (large) number of individuals.
- Measure gene expression for genes of interest.
- Assess the evidence that expression varies with genotype.
Linear additive model

Different alleles of a SNP may exhibit a dosage effect.
- Using the AA genotype as baseline, each copy of the B allele changes expression by a fixed amount.
- Implies a linear relationship between the mean gene expression and the number of B alleles.
- Estimating the change in expression due to the B allele to quantify the SNP’s contribution to gene expression.
Linear regression
\[Y = \beta_0 + \beta X + \varepsilon\]
- Y
- Response variable (here: vector of expression values for gene of interest).
- X
- Explanatory variable (here: vector of genotypes (coded as 0, 1, 2) for the SNP under consideration).
Linear regression
\[Y = \beta_0 + \beta X + \varepsilon\]
- \(\beta_0\)
- Intercept (here: mean expression for AA genotype).
- \(\beta\)
- Regression coefficient; the effect of X on the mean of Y (here: change in mean gene expression for each copy of the B allele).
- \(\varepsilon\)
- Residuals; the difference between observed values of \(Y\) and the estimated mean of \(Y|X\)
Linear regression – Assumptions
Residuals are
- independent
- normally distributed with mean 0 and constant variance
It is implied that
- There is a linear relationship between \(X\) and \(Y\).
- Values of \(Y\) for each value of \(X\) are normally distributed.
- There is only one source of variation not explained by \(X\).
Linear regression – Robustness
Independence of residuals
Lack of independence can produce misleading results.
What could cause this?
Linear regression – Robustness
Constant variance of residuals (homoskedacity)
Violation of this assumption will lead to incorrect p-values and confidence intervals.
Linear regression – Robustness
Normality
- Estimates and their confidence intervals and p-values are fairly robust.
- But beware of long tailed distributions.
- Prediction can become problematic (but we are not interested in that here).
Linear regression – Robustness
Linearity
If the true relationship between \(Y\) and \(X\) is non-linear conclusions may be misleading.
When might this occur with eQTL data?Linear regression – Robustness
Linearity
If the true relationship between \(Y\) and \(X\) is non-linear conclusions may be misleading.
When might this occur with eQTL data?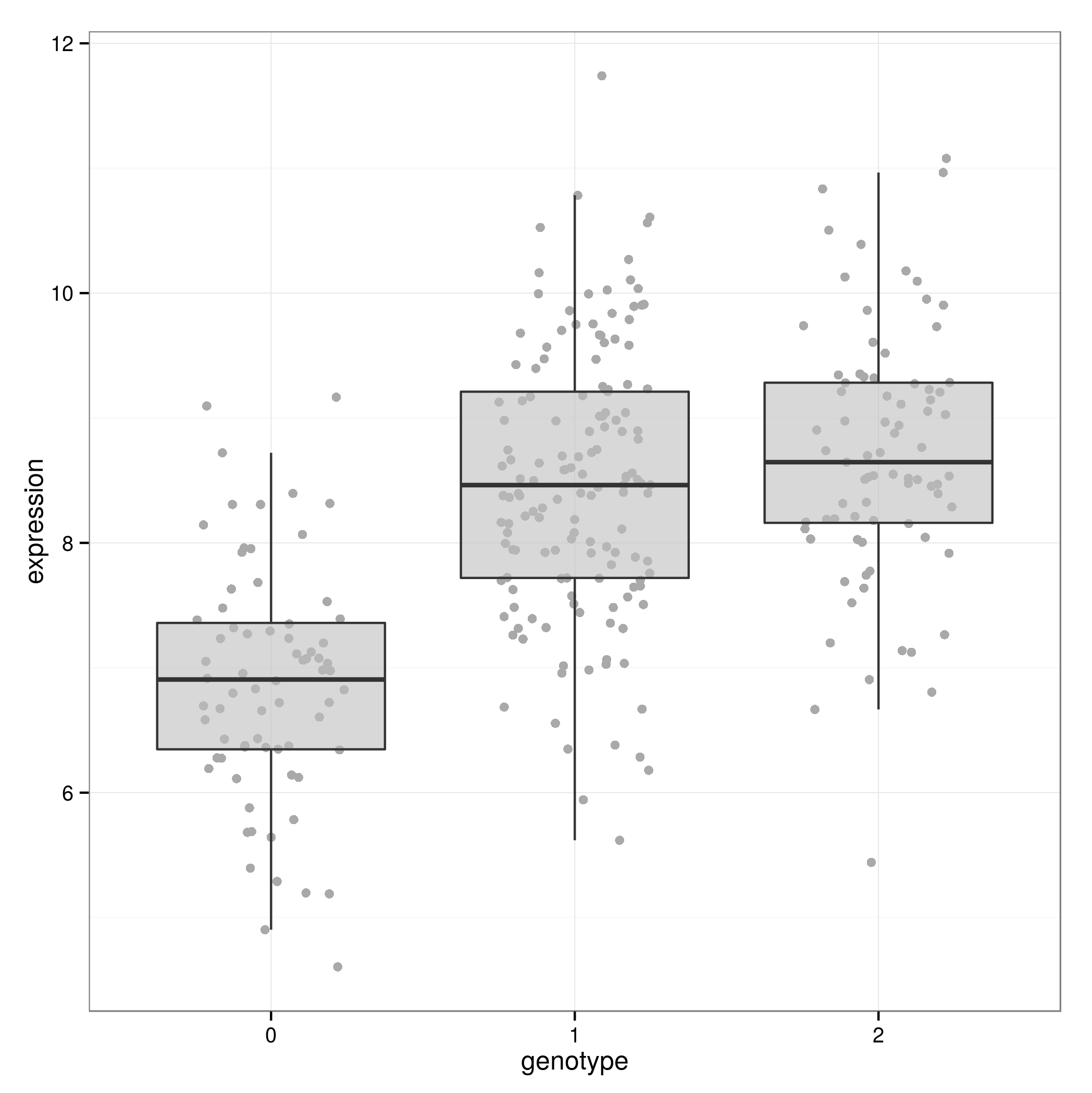
Hands-on : Simple linear regression
Data
- Genotypes
- /data/simulated/sim_genotypes.tab
- Gene expression
- /data/simulated/sim_expression1.tab
Warm-up
- Load the data into R.
- How is the data formatted?
- Determine minor allele frequencies.
Plotting the data
- Choose a SNP/gene pair (snp_1 / gene_1, snp_2 / gene_2, …)
- Create a plot showing gene expression by genotype for this pair.
Fitting a simple linear regression
- Fit a simple linear regression for the SNP/gene pair of your choice.
- Compute the 95% confidence interval for the genotype effect.
Examining the results
- Create diagnostic plots.
- Plot estimated genotype effects by minor allele frequency.
Interlude: p-values
- After estimating model coefficients we can test them for departure from 0.
- We are interested in identifying SNPs with non-zero coefficients.
- The strength of the evidence that the true coefficient is non-zero is often gauged by the resulting p-value.
- How should p-values be interpreted?
Detecting eQTL – Not that simple
Additional sources of variation
- Gene expression is subject to many sources of variation.
- A model with a single SNP as explanatory variable is unlikely to be sufficient.
Use multiple regression to obtain better estimates of SNP effects.
Multiple regression
\[Y = \beta_0 + \sum_{i=1}^n \beta_i X_i + \varepsilon\]
- Similar to simple linear regression but incorporates multiple explanatory variables.
- \(\beta_i\) are interpreted as “change of \(Y\) due to a unit change in \(X_i\) when all other explanatory variables are held constant”.
- Explanatory variables are assumed to be uncorrelated with each other.
Hands-on : Covariates
Data
- Genotypes
- /data/simulated/sim_genotypes.tab
- Gene expression
- /data/simulated/sim_expression2.tab
- Covariates
- /data/simulated/sim_covariates.tab
Plotting the data
Create a plot of gene expression by genotype for you SNP/gene pair of choice.
How does this compare to the plot from the previous exercise.
Simple linear regression
Repeat the simple linear regession analysis with these data.
- Fit a model for one of the SNP/gene pairs
- Create diagnostic plots
- Compute the confidence interval for the coefficient
How does this compare to the result from the previous analysis?
Multiple linear regression
- Use the first five variables contained in the covariates file as covariates in your model.
- Create diagnostic plots
- Compute the confidence interval for the coefficient
Covariates – Choose wisely
(Multi)-colinearity
If \(X_i\) and \(X_j\) are correlated the estimates of \(\beta_i\) and \(\beta_j\) will be biased. Several issues may occur:
All the variance in \(Y\) due to \(X_i\) and \(X_j\) is wholly attributed to one of the variables (say, \(X_i\)).
- Results in over estimation of \(\beta_i\) and under estimation of \(\beta_j\)
- Estimates for both \(\beta_i\) and \(\beta_j\) are too low because all of the variance is explained by the other variable that is held constant.
- No sensible interpretation for \(\beta_i\) and \(\beta_j\).
Model fitting may fail.
Dealing with colinearity
- Check correlation between explanatory variables.
- Check variance inflation factor (VIF).
- Choose only one from each group of correlated variables.
Only really need to worry about variables of interest for downstream analysis.
If only we knew – Covariates for real data
Limited information
- Real data comes with varying amounts of additional information.
- Sex and age are common
- May have detailed phenotyping data
- Some variables will have big impact on gene expression
- Won’t have information on all relevant variables.
Use the data
- We don’t have to know what the (non-genetic) sources of variation are as long as we can account for them.
- Dimensionality reduction techniques can identify major directions of variation from the data.
Principle component analysis
- Transforms the data into set of (linearly) uncorrelated variables (principle components).
- Principle components (PCs) are ordered by the proportion of variance they explain.
- Can reduce the dimensionality of a dataset by considering only the first \(k\) PCs.
How does that help us?
Accounting for unknown sources of variation
- The major sources of variation in gene expression data are (usually) not genetic.
- Can remove non-genetic sources of variation by including the3 first \(k\) PCs into the model.
Practical considerations
- Not obvious how many PCs to include in model.
- Need to look out for PCs that do correlate with genotype.
- Beware of data formatting issues.
- Data should be centred and scaled prior to PCA.
Hands-on : Dealing with real data
Data
- Gene expression
- /data/monocytes/expression/ifn_expression.tab.gz
- Genotypes
- /data/genotypes/genotypes.tab.gz
(downloaded earlier)
Subset of data published in
Fairfax, Humburg, Makino, et al.
Innate Immune Activity Conditions the Effect of Regulatory Variants upon Monocyte Gene Expression. Science (2014). doi:10.1126/science.1246949.
Brief look at the data
- Load the data and examine it
Interlude: Data processing
- These data have alredy been QC’d and processed.
- When dealing with raw data extensive QC is required for genotyping and expression data.
- Gene expression data also needs to be normalised across samples.
- Can be a lengthy process but is crucial for the quality of results.
PCA 101
- Compute principle components of gene expression data.
- Create a plot of the variances for the first 10 PCs.
- How much of the total variance is explained by the first 10 PCs?
Model fitting with PCA covariates
- Model the expression measured by probe 3710685 as a function of SNP rs4077515 and the first 10 PCs.
- Create a plot of gene expression by genotype with the effect of the PCs removed.
- How does this compare to the simple linear regression model for this SNP/gene pair.
Large scale eQTL analysis
Genome-wide analysis
- We don’t just want to analyse a single SNP/gene pair
- or even all SNP associations with a single gene
- We want to study all SNP/gene pairs.
- (but may restrict this to local associations).
This is computationally intensive and may be very time consuming.
Need to be clever about how we do this.
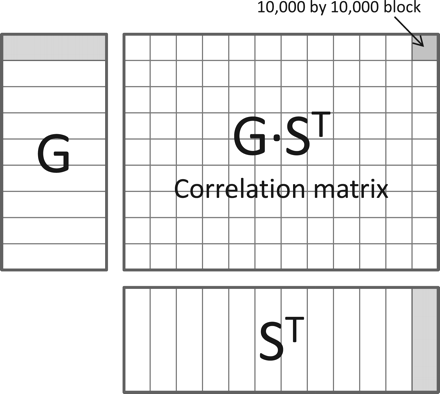
Fast model fitting with Matrix-eQTL
Matrix-eQTL strategies to reduce run time:- Required test statistic can be expressed in terms of the correlation between SNP and gene expression.
- Use efficient matrix multiplications to compute the correlations.
- Computing p-values is expensive, only do this for SNP/gene pairs that are sufficiently interesting.
Correcting for multiple testing
- A genome-wide analysis requires estimating millions of effect sizes.
- For an individual SNP/gene pair the p-value of the genotype coefficient may be a useful to gauge whether it is likely to be relevant.
- Interpreting p-values becomes more difficult if millions of them have to be considered.
False discovery rate
- Instead of considering each test individually try to control the number of tests that are incorrectly interpreted as indicating a true departure from the null hypothesis.
- Adjust p-values such that the resulting value is an estimate of the proportion of false positives obtained at a given threshold.
- Matrix-eQTL uses the Benjamini-Hochberg procedure.
Hands-on : Scaling it up
Data
- Gene expression
- /data/monocytes/expression/ifn_expression.tab.gz
- Genotypes
- /data/genotypes/genotypes.tab.gz
(downloaded earlier)
Annotation data
Located in /data/monocytes/annotation/
- snp_loc_hg19.tab
- Genomic location of SNPs.
- probe_loc_hg19.tab
- Genomic location of gene expression probes.
- probeAnnotations.tab
- Further annotations for gene expression probes, including associated gene symbols.
Running Matrix-eQTL
Use Matrix-eQTL to carry out a cis/trans eQTL analysis.
- Use a 1MB window around probes as local association region
- Use a p-value threshold of \(10^{-3}\) and \(10^{-5}\) for cis and trans associations respectively
- Repeat the analysis with the first 10 PC included as covariates. How do results differ?
Interpreting results
Still just a pile of data
- The initial analysis can produce a long list of SNP/gene associations.
- Without further analysis these are not particularly helpful.
- How can we identify particularly interesting results?
- How can we assign biological meaning to the list of associations?
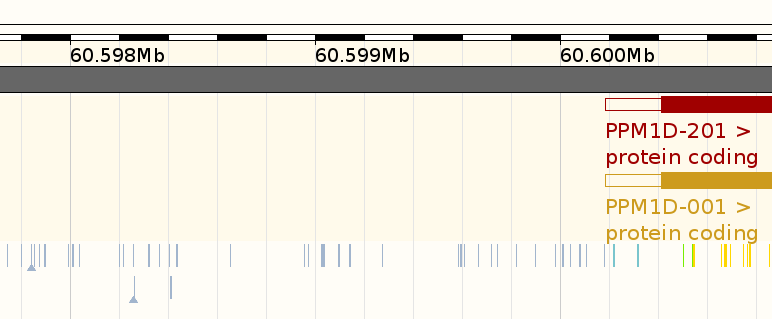
External data sources
- A lot of genomic datasets and derived annotations are available.
- Link SNPs to functional annotations using
- Visualise SNPs in genomic context, e.g. using UCSC genome browser
Summary
We talked about
- What eQTL are
- How linear models can be used to find them
- Complications that arise and possible approaches to deal with them
- How to carry out large scale eQTL analyses
- Some ideas of how to follow up on findings
We didn’t talk about
- Details of data QC
- The role of linkage disequilibrium in interpreting results
- Comparison of eQTL between different conditions (tissues, treatments, …)
- Other approaches to eQTL analysis (Bayesian, nonparametric)